Malaria remains one of Africa’s most persistent public health challenges, and early detection of disease vectors is critical to reducing transmission. A recent development in Madagascar highlights how artificial intelligence (AI) and citizen science are reshaping how malaria threats are identified and monitored across the continent.
According to a report by SciDev.Net, researchers used AI-powered image recognition tools alongside community participation to detect a potentially invasive malaria mosquito species, Anopheles stephensi, in Madagascar. This species is particularly concerning because it thrives in urban environments, breeding in water containers such as buckets, tyres, and tanks, settings where malaria transmission has traditionally been lower.
The detection was triggered when a citizen submitted a mosquito larva image via the NASA GLOBE Observer app. AI models trained on thousands of verified mosquito images classified the specimens with high confidence, demonstrating how digital tools can complement traditional entomological surveillance, especially in resource-limited settings.
For Africa, this approach has major implications. Conventional mosquito surveillance relies heavily on field trapping, microscopy, and laboratory confirmation, methods that are often expensive, slow, and unevenly distributed. AI-supported systems allow for rapid, scalable, and community-driven monitoring, improving early warning capacity and public health response.
At MyAfroDNA, we view this as part of a broader shift toward integrated genomic, digital, and environmental surveillance. Combining AI, biospecimen analysis, and genomic research can strengthen malaria control strategies, support vector mapping, and improve our understanding of how mosquito populations evolve and spread.
MyAfroDNA is actively seeking collaboration with researchers, public health institutions, universities, and malaria-focused organisations to support:
- Vector genomics and biospecimen research
- AI-assisted disease surveillance
- Data-driven malaria prevention strategies
Together, we can build African-led, evidence-based solutions for malaria research and control.
Reach out to collaborate with MyAfroDNA.
Source: SciDev.Net – AI and citizens detect invasive mosquito in Madagascar
https://www.scidev.net/sub-saharan-africa/news/ai-and-citizens-detect-invasive-mosquito-in-madagascar/

Public health surveillance has traditionally relied on clinics, hospitals, and laboratories to report cases of disease. But across much of Africa, this system captures only part of the picture. Many people do not seek care, cannot access testing, or are excluded from formal health systems altogether. As a result, outbreaks are often detected late, under-reported, or missed entirely.
A recent report in Nature highlights how wastewater surveillance is changing this reality, using sewage to track disease trends at the community level, even when clinical data is limited.
What is wastewater surveillance?
Wastewater surveillance involves testing sewage for genetic material from pathogens such as viruses and bacteria. When people are infected, traces of these pathogens are shed through bodily waste and end up in wastewater systems. By analysing these samples, scientists can estimate disease circulation across entire communities, without relying on individual testing or hospital visits.
In practical terms, one wastewater sample can represent thousands or even millions of people.
Lessons from South Africa
Researchers in South Africa demonstrated how wastewater monitoring can reveal disease trends that conventional surveillance fails to capture. By comparing viral signals in sewage with reported clinical cases, they found a significant gap between actual infections and officially recorded data.
In several instances, wastewater data showed rising infection levels weeks before clinics recorded an increase in cases. This early signal is especially important in lower-income or underserved areas, where testing rates tend to be lower, and outbreaks are more likely to go unnoticed.
The findings underscore a critical reality: absence of data does not mean absence of disease.
Beyond COVID-19
While wastewater surveillance gained prominence during the COVID-19 pandemic, its applications extend far beyond one virus. Researchers are now detecting genetic material linked to:
- Measles
- Mpox
- Influenza
- Hepatitis A and E
In some cases, these pathogens were identified in wastewater even when no clinical cases had been officially reported in the area. This positions wastewater surveillance as a powerful early-warning system for emerging and re-emerging diseases.
Why this matters for Africa
Wastewater surveillance offers several advantages that are particularly relevant in African contexts:
- Equity: It captures health data from entire communities, including people who are not represented in clinical systems.
- Cost-effectiveness: Monitoring a few sites can be more affordable than mass individual testing.
- Early detection: Public health authorities can respond sooner, potentially preventing wider outbreaks.
- Genomic insight: Sequencing wastewater samples allows scientists to track variants and pathogen evolution over time.
For regions facing resource constraints, these benefits can strengthen public health decision-making without placing additional burdens on individuals.
The challenges ahead
Despite its promise, wastewater surveillance is not without limitations. It requires:
- Laboratory infrastructure and sequencing capacity
- Skilled personnel for data analysis
- Sustainable funding and policy support
Encouragingly, continental initiatives, including efforts led by Africa CDC, are working toward integrating wastewater and environmental surveillance into broader disease monitoring systems.
Where MyAfroDNA fits in
At MyAfroDNA, we believe that genomic tools should serve real-world public health needs, especially in underrepresented regions. Wastewater surveillance demonstrates how genomics can move beyond laboratories and clinics to inform population-level health decisions.
As Africa continues to invest in biospecimen science, molecular testing, and genomic research, approaches like wastewater surveillance will be essential for building resilient, inclusive health systems.
Better data leads to better decisions and better outcomes for communities.

Recent advances published in the American Journal of Human Genetics highlight a pivotal shift in how scientists describe human populations in genetic studies — moving away from outdated racial terms toward more precise and meaningful descriptors like ancestry and ethnicity. In deepening our understanding of human genetic variation, language matters — both scientifically and ethically.
For decades, human genetics research sometimes relied on broad, socially loaded terms such as “race” or “Caucasian.” However, analyses tracking terms used in AJHG articles over time show a clear decline in the use of “race” and a rise in the use of labels like “African,” “European,” “Asian,” “ancestry,” and “ethnicity”. This shift reflects a growing recognition that continental labels and ancestry descriptors are more biologically and socially meaningful than simplistic racial categories.
This transition is not just semantic. Accurate terms improve how we design studies, interpret results and communicate findings — especially in genetics and genomic medicine, where variation is often deeply structured by geography, migration and population history rather than socially constructed groupings.
For a company like MyAfroDNA, this conversation underscores why African-centred genomics must be anchored in scientifically precise and culturally respectful language. Africa is the most genetically diverse continent on Earth, and understanding its genetic variation requires nuanced, context-specific frameworks rather than broad, imprecise categories.
By advocating for the use of ancestry and ethnicity labels rooted in deep genomic data — rather than traditional racial descriptors — the field is moving toward more accurate, inclusive and equitable genomics research. This evolution aligns with our mission: to enrich African genomic representation, empower informed interpretation, and advance science that reflects real human diversity.
Recent advances in genomics have revealed a new set of genes involved in regulating the glycosylation of immunoglobulin G (IgG)—a key antibody in the human immune system. Glycosylation, the process by which sugar molecules are attached to antibodies, directly affects immune signaling, inflammation, antibody stability, and therapeutic performance.
Traditionally, research has focused on enzymes that directly add or modify glycans. However, this new study highlights something more complex: many of the identified genes act as regulators, influencing glycosylation indirectly through gene expression, cellular pathways, and immune signaling networks. This finding reshapes how scientists understand immune variability and antibody behavior across individuals.
These insights are particularly important for biopharmaceutical development, where precise control of antibody glycosylation is critical for safety, efficacy, and consistency. Beyond therapeutics, IgG glycosylation patterns are also linked to autoimmune diseases, chronic inflammation, aging, and differential immune responses.
For Africa-focused genomics, the implications are profound. African populations harbor the highest genetic diversity globally, yet remain underrepresented in glycosylation and immunogenomics research. Without inclusive datasets, key regulatory variants may remain undiscovered, limiting the effectiveness of global precision medicine efforts.At MyAfroDNA, we believe that integrating African biospecimens and population-relevant genomic data is essential for advancing equitable immunology research and developing biotechnologies that work for everyone.
For full details and original findings, read the complete research article linked below.

A recent feature in Nature Africa highlights Tanzania’s remarkable progress in its decades-long battle against neglected tropical diseases (NTDs). Through coordinated mass drug administration, targeted surveillance, and strong community participation, the country has achieved a more than 75% reduction in key NTD infections, including lymphatic filariasis, trachoma, schistosomiasis and onchocerciasis.
This public-health milestone did not happen overnight. It is the result of consistent government investment, cross-sector partnerships, and the active involvement of local health workers who understand the unique cultural and environmental contexts of their communities. Village volunteers have played a crucial role in distributing medicines, tracking cases and ensuring that interventions reach even the most remote populations.The article also highlights that Tanzania’s approach is adaptable. For example, as prevalence levels dropped, the country shifted from broad mass treatment campaigns to more precise surveillance systems, using community data to identify hotspots and allocate resources more efficiently.
This transition demonstrates the power of combining field-driven insights with data-led public-health decision-making.For a company like MyAfroDNA, Tanzania’s progress offers a compelling model for African-led health innovation. It reinforces an essential truth: African health systems are not passive recipients of external interventions. They are active, evidence-driven and capable of delivering measurable, population-level impact when local leadership is centred.These lessons are highly relevant to our molecular-testing, biobanking and biospecimen work.
Sustainable impact in genomics requires:partnerships that elevate community expertise,programmes built on local context rather than imported assumptions, andlong-term models that integrate scientific data with lived realities.Tanzania’s success in reducing NTD burdens is more than a public-health victory. It illustrates what becomes possible when data, community participation and local ownership intersect. As MyAfroDNA advances its African-centred genomics agenda, this achievement reminds us of the responsibility — and opportunity — in supporting African science, strengthening community-driven systems and amplifying homegrown impact.
Read the full article here
Africa is achieving one of its most remarkable public-health milestones in recent memory: the continent has reported a 28% drop in tuberculosis (TB) incidence and a 46 % decline in TB-related deaths, figures that surpass global targets and highlight a critical shift in disease control capacity.
This progress comes despite persistent challenges: constrained funding, limited infrastructure, and complex cross-border dynamics across Africa’s health systems. Yet it also underscores an important truth: when scientific tools, coordinated policy, and local leadership align, even historically burdensome diseases can be brought under control. For organisations working in molecular diagnostics and biospecimen services like yours, this achievement offers both inspiration and context.
At MyAfroDNA, this success story reinforces our core mission: making high-quality molecular testing accessible across Africa. As TB incidence and mortality decline, the demand for accurate diagnostics, pathogen genomics, and robust biosamples becomes ever more critical. Lower TB rates elevate the importance of refined testing services that can support surveillance, differential diagnosis, and broader public-health tracking. Moreover, biobanks and specimen-sharing frameworks become integral in documenting evolving pathogens and designing targeted interventions.
In practical terms, this means we must scale our molecular-testing infrastructure, strengthen our collaboration with health systems, and ensure our biospecimens meet rigorous standards. By doing so, we contribute to a future where African health systems are not just battling disease but staying ahead of it. The continent’s TB victory is not just one battle won; it’s a warning light for the next frontier of diagnostics, genomics, and equitable health innovation.
Read the full article here: Africa exceeds global tuberculosis targets, despite funding squeeze
Explore how MyAfroDNA’s molecular-testing and DNA-diagnostic services are helping build Africa’s health-tech future. Learn more about our services to learn more and partner with us.

The official launch of the African Medicines Agency (AMA) marks a historic turning point for health innovation and pharmaceutical regulation across the continent. Established to streamline the approval and monitoring of medical products, the AMA aims to unify Africa’s fragmented regulatory systems and strengthen the fight against counterfeit or substandard medicines.
According to a recent Nature article, the creation of this central regulatory body promises to improve access to safe, effective, and affordable healthcare solutions. It also paves the way for biotech growth, ensuring that diagnostic tools, vaccines, and molecular testing technologies meet shared standards across African nations. For biotech companies, researchers, and laboratories like MyAfroDNA, this move signals greater collaboration and credibility within global scientific frameworks.
However, the article also points out the challenges ahead. Differences in national policies, limited funding, and uneven political commitment could slow the pace of implementation. To truly succeed, the AMA will require consistent investment in local expertise, capacity building, and transparent governance.
At MyAfroDNA, we see this milestone as an invitation to deepen our role in Africa’s biotechnology ecosystem. As a molecular testing, DNA diagnostics, and biobanking company, our work depends on strong regulatory systems that protect patients and ensure scientific integrity. The AMA’s commitment to standardized, ethical practices aligns perfectly with our mission to make accurate, reliable testing accessible across Africa.
Further reading: What the launch of the African Medicines Agency means for drug and health regulation – Nature (2025)
Explore how MyAfroDNA’s molecular testing and DNA diagnostic services are setting new standards for biospecimen quality and public health in Africa. Visit www.myafrodna.com to learn how we’re advancing Africa’s biotech future, one test at a time.
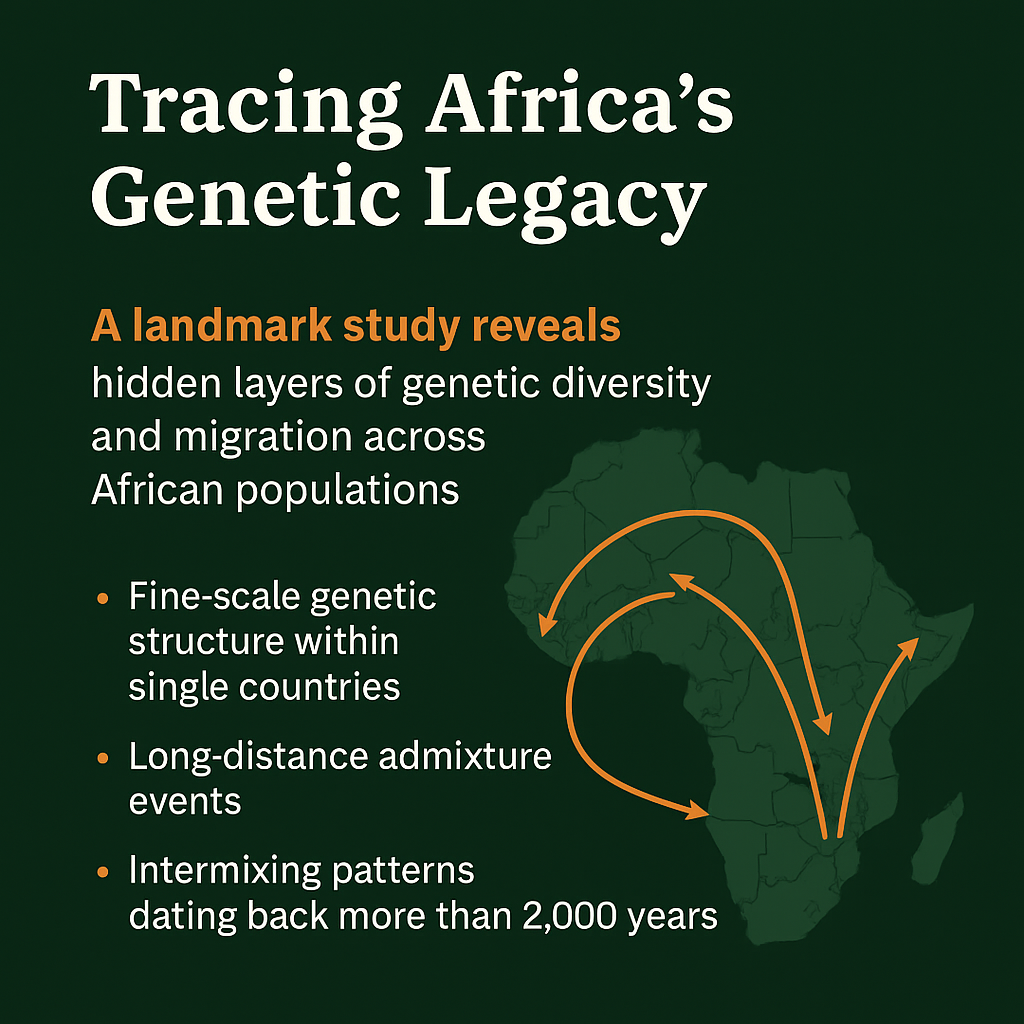
A landmark study led by researchers from University College London (UCL) and published in Science Advances reveals a previously hidden depth of genetic diversity, migration, and admixture within African populations. Titled “Dense sampling of ethnic groups within African countries reveals fine-scale genetic structure and extensive historical admixture,” the research analysed 1,333 genomes from over 150 ethnic groups across Cameroon, Ghana, Nigeria, Sudan, and the Republic of the Congo.
The findings challenge the often simplistic narratives about African genetic history. The study uncovered fine-scale genetic structure within single countries, showing that even neighbouring ethnic groups may carry distinct ancestral lineages. For instance, western Cameroonian groups exhibit unique ancestry signatures reflecting the region’s long history of local kingdoms and cultural interactions.
Researchers also traced long-distance admixture events, linking populations in northern Cameroon and Sudan with distant groups, suggesting centuries of movement through trade, migration, and empire expansion. In Ghana and Nigeria, they detected intermixing patterns dating back more than 2,000 years, likely connected to shifts in climate and vegetation that encouraged population mobility and contact.
Beyond uncovering these complex patterns, the study highlights an essential truth: Africa’s genomic diversity cannot be fully understood through limited or external data. It underscores the urgency of expanding and diversifying African genomic datasets to ensure equitable representation in global genetics research.
For MyAfroDNA, this research reaffirms our mission to strengthen African-centric molecular testing and biospecimen sourcing for both research and precision medicine. Understanding these fine-scale patterns helps scientists interpret genetic variation more accurately, improving ancestry insights and health-related findings for African communities.
As Africa continues to shape the global genomic landscape, studies like this remind us that every region, community, and ancestry carries its own genetic legacy, one that deserves to be studied, respected, and represented on its own terms.
Read the full research here.
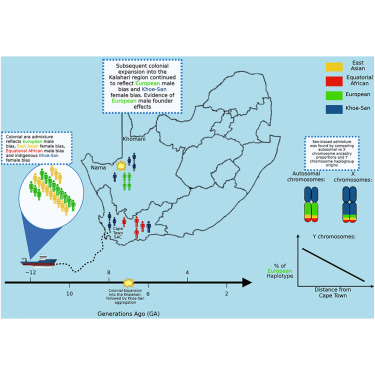
A new study published in The American Journal of Human Genetics reveals how centuries of colonial expansion and the Indian Ocean slave trade shaped South Africa’s genetic landscape, leaving a profound legacy of sex-biased admixture.
Researchers analyzed genetic data from over 1,400 individuals across South Africa to understand how migration and displacement transformed Indigenous communities. The findings show that European male settlers contributed disproportionately to genetic lineages, while Khoe-San women and enslaved women from South and Southeast Asia made major contributions to the maternal gene pool.
Interestingly, while genetic mixing around the Cape was continuous, northern Khoe-San communities experienced a single pulse of European admixture about six to eight generations ago. The Nama people showed unique founder effects, with about 15% of Y-chromosome lineages tracing back to Asia, reflecting the deep genetic impact of forced migrations during colonial times.
This research highlights how genomics can uncover stories of resilience and connection, offering new insight into Africa’s intertwined histories of movement, survival, and identity.
At MyAfroDNA, we are committed to advancing genomics research by providing high-quality African biospecimens and molecular testing services that help decode Africa’s diverse genetic heritage.
Click here for further reading.

