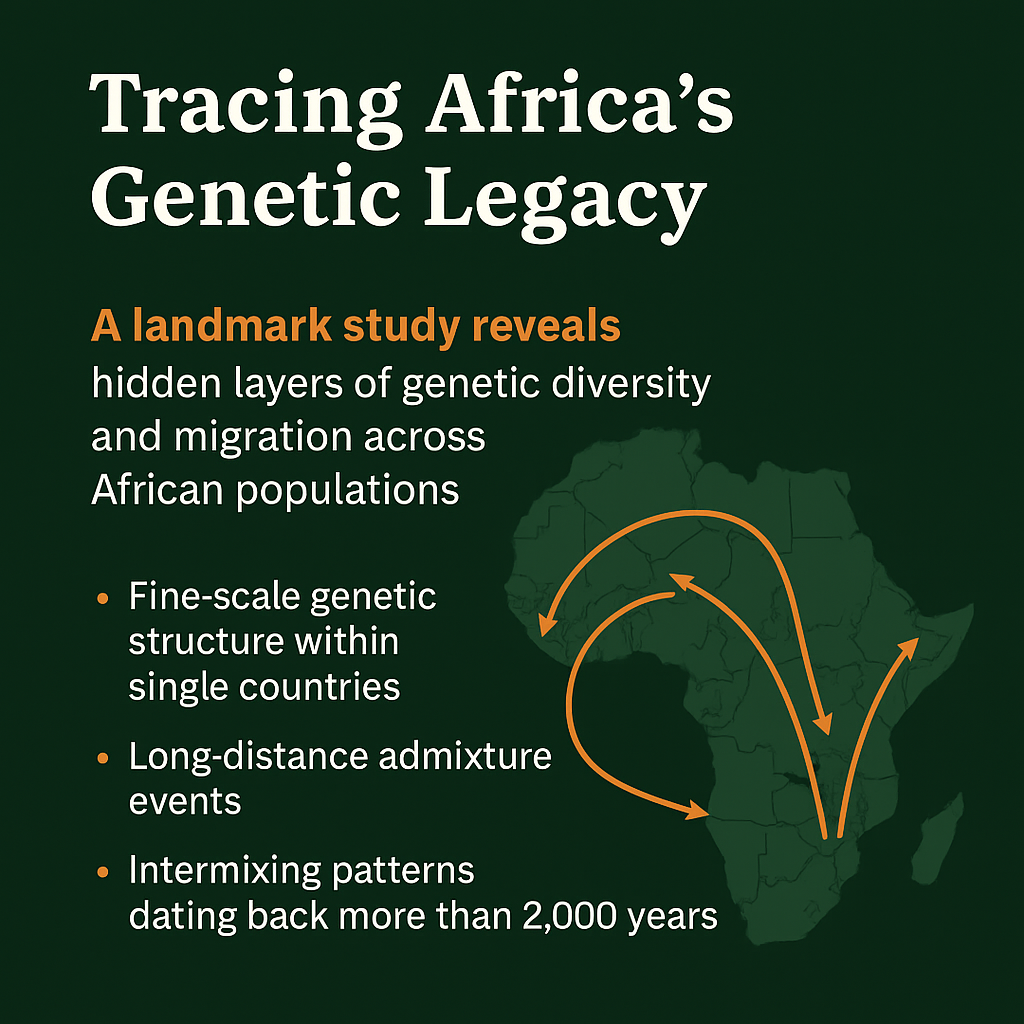
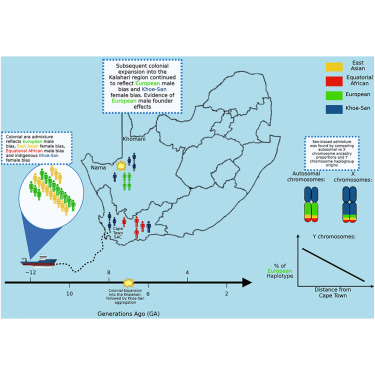
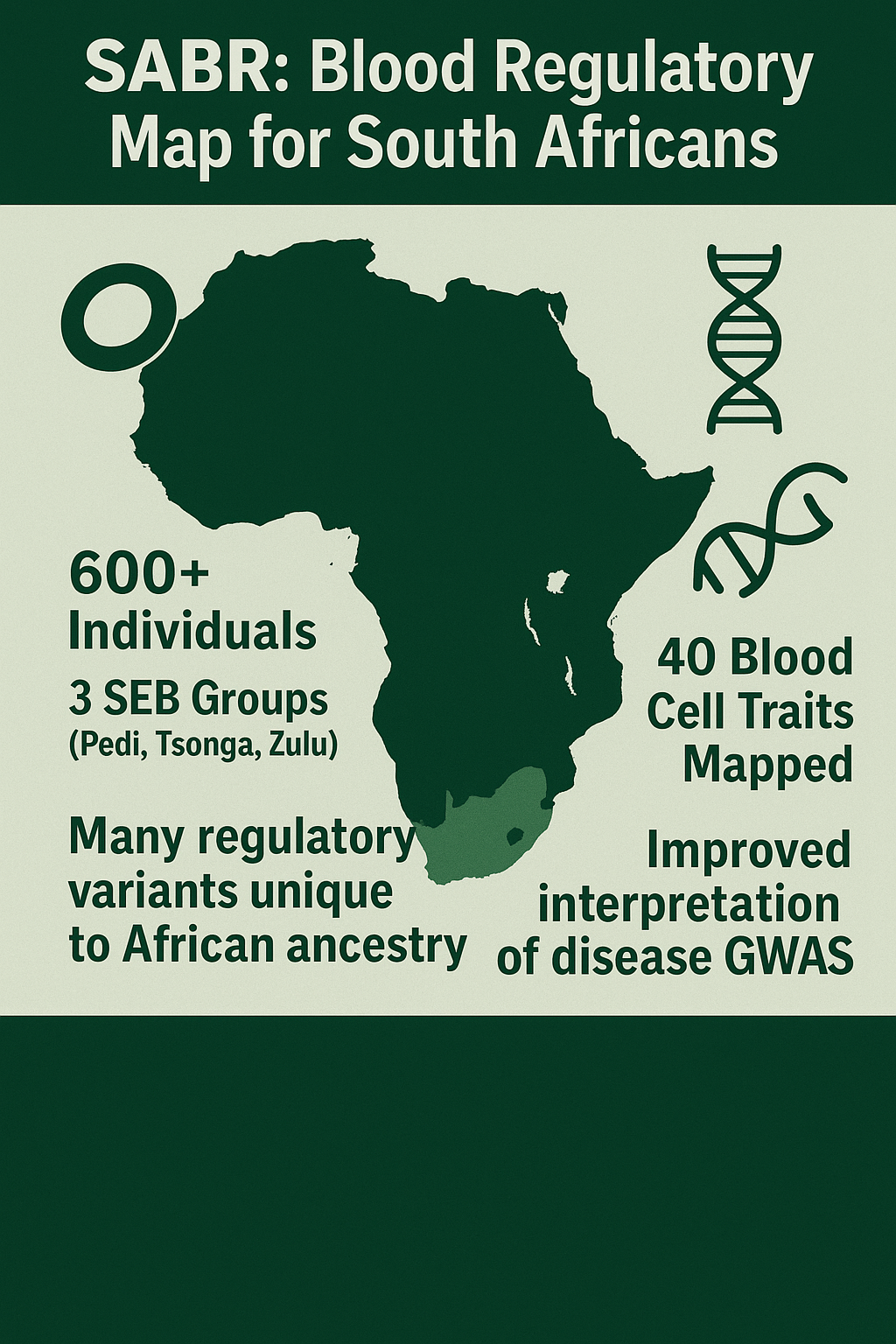
A new study published in Nature projects that climate change, especially extreme weather events could lead to a substantial increase in malaria cases and deaths across Africa over the next 25 years.
Using 25 years of climate, health, socioeconomic, and disease control data, researchers estimate that climate change could contribute to up to 123 million additional malaria cases and more than 500,000 additional deaths between 2024 and 2050 if current control strategies remain unchanged.
While many studies have focused on how temperature and rainfall affect mosquito ecology, this research shows that extreme weather such as floods and cyclones — will be the main driver of increased risk, disrupting housing, health services, prevention programs, and access to treatment. These disruptions may account for the majority of additional cases and deaths in the coming decades.
Most of the projected increases will occur in regions where malaria is already endemic rather than in entirely new areas, underscoring the vulnerability of existing control systems to climate shocks. These findings highlight a profound challenge: climate change doesn’t just reshape environments, it can undermine disease control infrastructures, particularly where health systems are already fragile.
For genomic scientists and public health researchers, this study reinforces the urgency of climate-resilient malaria strategies that integrate environmental forecasting, vector surveillance, health system preparedness, and genomic research to track parasite and vector evolution under changing conditions.
At MyAfroDNA, we are committed to supporting collaborative research that advances:Climate-informed malaria surveillance Integrated vector genomics and environmental data systemsCommunity-embedded biospecimen collection and analysisData-driven intervention and control strategiesIf you are a researcher, institution, or organization working on malaria, climate health, genomics, or vector-borne disease research, we invite you to partner with MyAfroDNA. Together, we can develop solutions that are tailored to Africa’s climate and health needs.