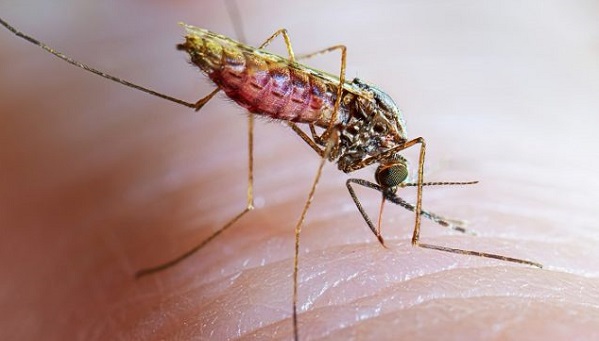
Malaria remains one of Africa’s most persistent public health challenges, and early detection of disease vectors is critical to reducing transmission. A recent development in Madagascar highlights how artificial intelligence (AI) and citizen science are reshaping how malaria threats are identified and monitored across the continent.
According to a report by SciDev.Net, researchers used AI-powered image recognition tools alongside community participation to detect a potentially invasive malaria mosquito species, Anopheles stephensi, in Madagascar. This species is particularly concerning because it thrives in urban environments, breeding in water containers such as buckets, tyres, and tanks, settings where malaria transmission has traditionally been lower.
The detection was triggered when a citizen submitted a mosquito larva image via the NASA GLOBE Observer app. AI models trained on thousands of verified mosquito images classified the specimens with high confidence, demonstrating how digital tools can complement traditional entomological surveillance, especially in resource-limited settings.
For Africa, this approach has major implications. Conventional mosquito surveillance relies heavily on field trapping, microscopy, and laboratory confirmation, methods that are often expensive, slow, and unevenly distributed. AI-supported systems allow for rapid, scalable, and community-driven monitoring, improving early warning capacity and public health response.
At MyAfroDNA, we view this as part of a broader shift toward integrated genomic, digital, and environmental surveillance. Combining AI, biospecimen analysis, and genomic research can strengthen malaria control strategies, support vector mapping, and improve our understanding of how mosquito populations evolve and spread.
MyAfroDNA is actively seeking collaboration with researchers, public health institutions, universities, and malaria-focused organisations to support:
- Vector genomics and biospecimen research
- AI-assisted disease surveillance
- Data-driven malaria prevention strategies
Together, we can build African-led, evidence-based solutions for malaria research and control.
Reach out to collaborate with MyAfroDNA.
Source: SciDev.Net – AI and citizens detect invasive mosquito in Madagascar
https://www.scidev.net/sub-saharan-africa/news/ai-and-citizens-detect-invasive-mosquito-in-madagascar/